
MtDNA haplogroups
All mtDNA haplogroups found outside of Africa are descendants of either haplogroup N or its sibling haplogroup M. M and N are the signature haplogroups that define the out of Africa migration and the subsequent spread to rest of the world. The global distribution of haplogroups N and M, indicates that very likely, there was one particularly major prehistoric migration of humans out of Africa, and both N and M were part of the same colonization process.
An enormous haplogroup spanning many continents, the macro-haplogroup N, like its sibling M, is a descendant of haplogroup L3. There is widespread agreement in the scientific community concerning the African ancestry of haplogroup L3 (haplogroup N’s parent clade). However, whether or not the mutations which define haplogroup N itself first occurred within Asia or Africa has been a subject for ongoing discussion and study.
Haplogroup N is derived from the ancestral L3 haplotype that represents the ‘Out of Africa’ migration. Haplogroup N is the ancestral haplogroup to almost all European and Oceanian haplogroups in addition to many Asian and Amerindian ones. It is believed to have arisen at a similar time to haplogroup M.
MtDNA haplogroup R is a very extended mitochondrial DNA (mtDNA) haplogroup and is the most common macro-haplogroup in West Eurasia. It is a descendant of macro-haplogroup N. Among its descendant haplogroups are B, U (and thus K), F, R0 (and thus HV, H, and V), and the ancestral haplogroup of J and T. As of June 2009, the most recent study dates the origin of haplogroup R to 66.8kya with a 95% confidence interval of 52.6-81kya.
South Asia lies on the way of earliest dispersals from Africa and is therefore a valuable well of knowledge on early human migration. The analysis of the indigenous haplogroup R lineages in India points to a common first spread of the root haplotypes of M, N, and R along the southern route some 60–70 kya.
Haplogroup R has wide diversity and antiquity among varied ethnic status and different language families in South Asia. In Indian western region among the castes and southern region among the tribes show higher haplogroup diversity than the other regions, possibly suggesting their autochthonous status.
MtDNA Haplogroup U descends from a woman in the mtDNA haplogroup R branch of the phylogenetic tree, who lived around 55,000 years ago. It is widely distributed across Western Eurasia, North Africa, and South Asia.
Subclades such as Haplogroup U6, are also found at moderate to low frequencies in the Northwest and East Africa, due to a back migration from Asia around 27,000 years ago. In spite of the highest diversity of Iberian U6, Maca-Meyer argues for an Near East origin of this clade based on the highest diversity of subclade U6a in that region, where it would have arrived from West Asia. She estimates the age of U6 between 25,000 and 66,000 years BP.
Controversy surrounds the existence of mitochondrial DNA haplogroup N in Africa. Some researchers have suggested that haplogroups N probably originated in Africa (Quintana-Murci et al, 1999; Sun el al,2005). Quintana-Murci et al (1999) has suggested that haplogroup N probably originated in Ethiopia before the out of Africa migration.
Other researchers believe that haplogroup Nin Africa is the result of a back migration. The cra-niofacial and molecular evidence does not supportthis conclusion. The molecular evidence indicatesthat haplogroup N is found across Africa from Eastto West on into India where it was deposited by Dravidian speakers (Winters, 2007, 2008).
Archaeogenetics is the use of genetics, ar-chaeology and linguistics to explain and discuss theorigin and spread of Homo sapiens. Using thismethodology we can gain valuable insight into hu-man history and population movements in prehis-toric times.
In this paper we will examine the spread of haplogroup N from Africa to Eurasia. It will either support an African origin or, back migration for the presence of the N haplogroup in Africa
Origin and Spread of Haplogroup N
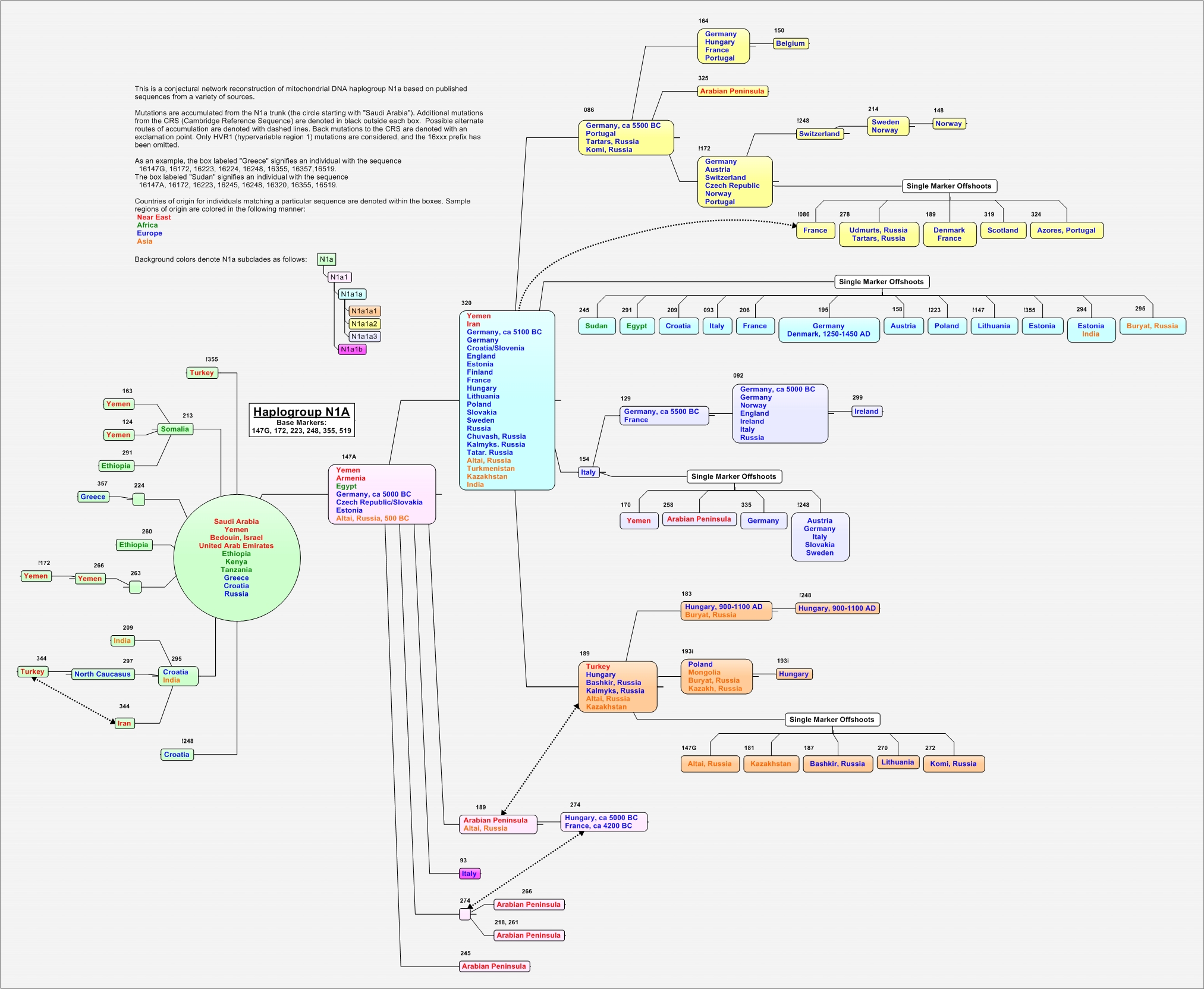
MtDNA haplogroup N1a
The current human mitochondrial (mtDNA) phylogeny does not equally represent all human populations but is biased in favour of representatives originally from north and central Europe. This especially affects the phylogeny of some uncommon West Eurasian haplogroups, including I and W, whose southern European and Near Eastern components are very poorly represented, suggesting that extensive hidden phylogenetic substructure remains to be uncovered.
This study expanded and re-analysed the available datasets of I and W complete mtDNA genomes, reaching a comprehensive 419 mitogenomes, and searched for precise correlations between the ages and geographical distributions of their numerous newly identified subclades with events of human dispersal which contributed to the genetic formation of modern Europeans.
Our results showed that haplogroups I (within N1a1b) and W originated in the Near East during the Last Glacial Maximum or pre-warming period (the period of gradual warming between the end of the LGM, ~19 ky ago, and the beginning of the first main warming phase, ~15 ky ago) and, like the much more common haplogroups J and T, may have been involved in Late Glacial expansions starting from the Near East.
Thus our data contribute to a better definition of the Late and postglacial re-peopling of Europe, providing further evidence for the scenario that major population expansions started after the Last Glacial Maximum but before Neolithic times, but also evidencing traces of diffusion events in several I and W subclades dating to the European Neolithic and restricted to Europe.
Haplogroup N1a
Haplogroup N1a is widely distributed throughout Eurasia and Eastern Africa and is divided into the European/Central Asian and African/South Asian branches based on specific genetic markers.
N1a originated in the Near East 12,000 to 32,000 years ago. Specifically, the Arabian Peninsula is postulated as the geographic origin of N1a. This supposition is based on the relatively high frequency and genetic diversity of N1a in modern populations of the peninsula. Exact origins and migration patterns of this haplogroup are still subject of some debate.
The tree of N1a has two distinct branches: Africa/South Asia and Europe with a Central Asian subcluster. However, the African branch has members in southern Europe, and the European branch has members in Egypt and the Near East. The Africa/South Asia branch is characterized by the 16147G mutation, whereas the European branch is characterized by 16147A, 3336, and 16320. The Central Asian subcluster is an offshoot of the European branch that is characterized by marker 16189.
Subclade N1a1 is associated with mutation 16147A. Palanichamy calculates N1a1 to have emerged between 8900 to 22400 YBP. Subclade N1a1a is denoted by marker 16320, and is therefore associated with the “European” N1a branch. Petraglia estimates that N1a1a arose between 11000 to 25000 YBP.
Two main competing scenarios exist for the spread of the Neolithic from the Near East to Europe: Demic diffusion (in which farming is brought by farmers) vs. Cultural diffusion (in which farming is spread by the passage of ideas).
N1a became particularly prominent in this debate when a team led by Wolfgang Haak analyzed skeletons from Linear Pottery Culture, credited with the first farming communities in Central Europe, marking the beginning of Neolithic Europe in the region some 7500 years ago.
As of 2010, mitochondrial DNA analysis has been conducted on 42 specimens from five locations. Seven of the 42 specimens were found to be members of haplogroup N1a.
A separate study analyzed 22 skeletons from European hunter-gatherer sites dated 13400-2300 BC. Most of these remains were members of Haplogroup U, which was not found in any of the Linear Pottery Culture sites. Conversely, N1a was not identified in any of the hunter-gatherer fossils, indicating a genetic distinction between early European farmers and late European hunter-gatherers.
While no modern population is a close match to the LBK findings, the authors claim that the Linear Pottery population is most closely affiliated with modern Near East populations. Given this affiliation and the group’s distinctiveness from hunter-gatherers, Haak’s team concludes that “the transition to farming in central Europe was accompanied by a substantial influx of people from outside the region.”
However, they note that haplogroup frequencies in modern Europeans are substantially different from early farming and late hunter-gatherer populations. This indicates that “the diversity observed today cannot be explained by admixture between hunter-gatherers and early farmers alone” and that “major demographic events continued to take place in Europe after the early Neolithic.”
Critics of these studies claim that the LBK N1a specimens could have derived from local communities established in Europe before the introduction of farming. Ammerman’s team voiced concern due to some of the LBK specimens coming from communities several hundred years after farming was first established in the region; a rebuttal was given.
In 2010, researchers led by Palanichamy conducted a genetic and phylogeographic analysis of N1a. Based on the results, they conclude that some of the LBK samples were indigenous to Europe while others may have resulted from ‘leapfrog’ colonization.
Deguilloux’s team agreed with Haak’s conclusion on a genetic discontinuity between ancient and modern Europeans. However, they consider demic diffusion, cultural diffusion, and long-distance matrimonial exchanges all equally plausible explanations for the current genetic findings.
Seven of 42 skeletons from Linear Pottery Culture sites were found to be members of the N1a haplogroup (see Neolithic European section). N1a was also identified in remains from a 6200 year-old megalithic long mound near Prissé-la-Charrière, France. A 2500 year old fossil of a Scytho-Siberian in the Altai Republic, easternmost representative of the Scythians, was found to be a member of N1a1.
A study of a 10th and 11th century Hungarians found that N1a1a1 was present in high-status individuals but absent from commoners. One of thirteen skeletons analyzed from a medieval cemetery dated 1250-1450 AD in Denmark was found to be a member of subclade N1a1a.
Mitochondrial haplogroup N1a phylogeography, with implication to the origin of European farmers
Danubian Neolithic ancient mtDNA N1a is (mostly) European by origin
Dienekes’ Anthropology Blog: Origin of Neolithic N1a
Dienekes’ Anthropology Blog: N1a in Brahmnins
Mitochondrial haplogroup N1a phylogeography
Ancient DNA From the First European Farmers in 7500-Year-Old Neolithic Sites
Origin and Spread of Haplogroup N
Haplogroup N1a (mtDNA) – Familypedia
Family Tree DNA – The Haplogroup N mtDNA Study
Filed under: Uncategorized
