



Most parsimonious phylogeny of YAP
(Underhill and Kivisild 2007)
Haplogroup E (Y-DNA)
The Worsening Biomedical Crisis
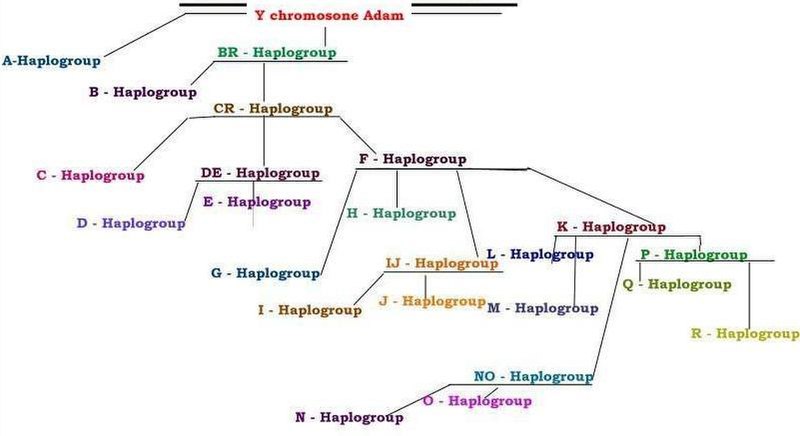
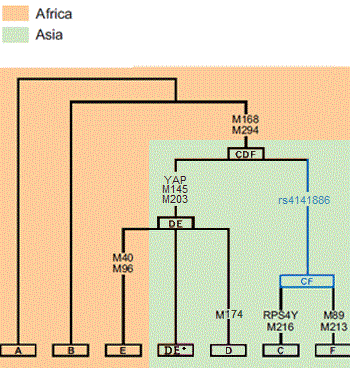
In human genetics, Haplogroup DE is a human Y-chromosome DNA haplogroup. It is defined by the single nucleotide polymorphism (SNP) mutations, or UEPs, M1(YAP), M145(P205), M203, P144, P153, P165, P167, P183.
Haplogroup DE is often referred to by the most well-known unique event polymorphism (UEP) which defines it, the Y-chromosome Alu Polymorphism (YAP). The YAP mutation was caused when a strand of DNA called Alu, which copies itself, inserted a copy into the Y chromosome.
A Y chromosome that has the YAP mutation is called YAP-positive (YAP+), and a Y chromosome that does not have the YAP mutation is labeled YAP-negative (YAP-).
Haplogroup DE is an estimated 65,000 years old.
The majority of DE male lines can be categorized as being in either Haplogroup D (Y-DNA), which likely originated in Asia, the only place where it has been found, or haplogroup E, which is believed to have originated in East Africa or the Near East. The remainder are said to be in the paragroup DE*, confirmed cases of which are extremely rare.
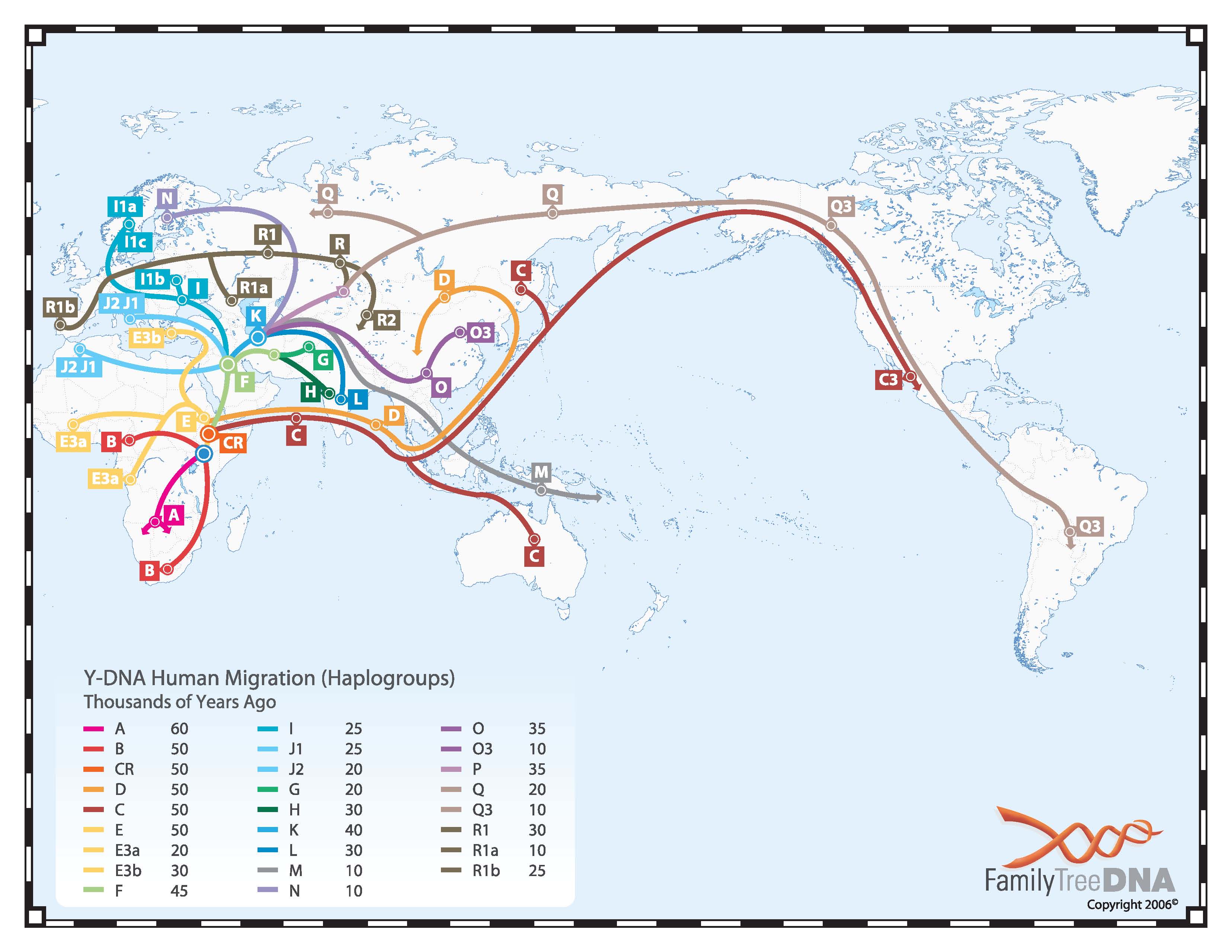
Haplogroup DE is found in Africa (Haplogroups E and DE*) and East Asia (Haplogroups D and DE* and E*) but is largely absent in between these two regions.
The presence of DE across widely separated regions has confounded investigators trying to reconstruct the migration of humans from Africa to Asia. At some time, there was an extinction of DE lineages in West, South and Central Asia.
Autochthonous DE lineages are absent in India, an important region in the dispersal of humans in Asia. However DE lineages have been detected in relict populations of the Andaman Islands.
Underhill et al. 2007, suggest the possibility that deleterious mutations in some DE carriers may explain the extinction of DE lineages in India.
In human genetics, Haplogroup D-M174 is a Y-chromosome haplogroup. Both D-M174 and E lineages also exhibit the single-nucleotide polymorphism M168 which is present in all Y-chromosome haplogroups except A and B, as well as the YAP unique-event polymorphism, which is unique to Haplogroup DE.
Haplogroup D-M174 is believed to have originated in Asia some 60,000 years before present. While haplogroup D-M174 along with haplogroup E contains the distinctive YAP polymorphism (which indicates their common ancestry), no haplogroup D-M174 chromosomes have been found anywhere outside of Asia.
Like haplogroup C, D-M174 is believed to represent the Great Coastal Migration along southern Asia, from Arabia to Southeast Asia and thence northward to populate East Asia. It is found today at high frequency among populations in Tibet, the Japanese Archipelago, and the Andaman Islands, though curiously not in India.
The Ainu of Japan and the Jarawa and Onge of the Andaman Islands are notable for possessing almost exclusively Haplogroup D-M174 chromosomes, although Haplogroup C-M217 chromosomes also have been found in 15% (3/20) of sampled Ainu males.
Haplogroup D-M174 chromosomes are also found at low to moderate frequencies among populations of Central Asia and northern East Asia as well as the Han and Miao–Yao peoples of China and among several minority populations of Sichuan and Yunnan that speak Tibeto-Burman languages and reside in close proximity to the Tibetans.
Unlike haplogroup C-M217, Haplogroup D-M174 is not found in the New World; it is not present in any modern Native American (North, Central or South) populations. While it is possible that it traveled to the New World like Haplogroup C-M217, those lineages apparently became extinct.
Haplogroup D-M174 is also remarkable for its rather extreme geographic differentiation, with a distinct subset of Haplogroup D-M174 chromosomes being found exclusively in each of the populations that contains a large percentage of individuals whose Y-chromosomes belong to Haplogroup D-M174:
Haplogroup D-M15 among the Tibetans (as well as among the mainland East Asian populations that display very low frequencies of Haplogroup D-M174 Y-chromosomes), Haplogroup D-M55 among the various populations of the Japanese Archipelago, Haplogroup D-P99 among the inhabitants of Tibet, Tajikistan and other parts of mountainous southern Central Asia, and paragroup D-M174 without tested positive subclades (probably another monophyletic branch of Haplogroup D) among the Andaman Islanders.
Another type (or types) of paragroup D-M174 without tested positive subclades is found at a very low frequency among the Turkic and Mongolic populations of Central Asia, amounting to no more than 1% in total.
This apparently ancient diversification of Haplogroup D-M174 suggests that it may perhaps be better characterized as a “super-haplogroup” or “macro-haplogroup.” In one study, the frequency of Haplogroup D-M174 without tested positive subclades found among Thais was 10%.
The Haplogroup D-M174 Y-chromosomes that are found among populations of the Japanese Archipelago are particularly distinctive, bearing a complex of at least five individual mutations along an internal branch of the Haplogroup D-M174 phylogeny, thus distinguishing them clearly from the Haplogroup D-M174 chromosomes that are found among the Tibetans and Andaman Islanders and providing evidence that Y-chromosome Haplogroup D-M5 was the modal haplogroup in the ancestral population that developed the prehistoric Jōmon culture in the Japanese islands.
In human genetics, Haplogroup E-M96 is a human Y-chromosome DNA haplogroup. Haplogroup E-M96 is one of the two main branches of the older Haplogroup DE, the other main branch being haplogroup D. The E-M96 clade is divided into two subclades: The more common E-P147 and the less common E-M75.
Underhill (2001) proposed that haplogroup E may have arisen in East Africa. Some authors as Chandrasekar (2007), continue to accept the earlier position of Hammer (1997) that Haplogroup E may have originated in Asia, given that:
E is a clade of Haplogroup DE, with the other major clade, haplogroup D, being East Asian.
DE is a clade within M168 with the other two major clades, C and F, considered to have a Eurasian origin.
However, several discoveries made since the Hammer articles are thought to make an Asian origin less likely:
Underhill and Kivisild (2007) demonstrated that C and F have a common ancestor meaning that DE has only one sibling which is non-African.
DE* is found in both Asia and Africa, meaning that not only one, but several siblings of D are found in Asia and Africa.
Karafet (2008), in which Hammer is a co-author, significantly rearranged time estimates leading to “new interpretations on the geographical origin of ancient sub-clades”.
Amongst other things this article proposed a much older age for haplogroup E-M96 than had been considered previously, giving it a similar age to Haplogroup D, and DE itself, meaning that there is no longer any strong reason to see it as an offshoot of DE which must have happened long after DE came into existence and had entered Asia.
Most members of haplogroup E-M96 belong to one of its identified subclades, and the E-M96 (xE-P147,xE-M75) lineage is rare.
E1a and E-M75 are found almost exclusively in Africa. By looking at the major subclade frequencies, five broad regions of Africa can be defined: East, Central, North, Southern and West.
The division can be distinguished by the prevalence of E-V38 in East, Central, Southern and West Africa, E-M78 in East Africa and E-M81 in North Africa.
E-V38 is the most prevalent subclade of E in Africa. It is observed at high frequencies in all African regions except the northernmost and easternmost portions of the continent.
E-M243 (especially its subclades M78 and M81) is found at high frequencies in North East Africa and North Africa and is the only subclade that is found in Europe and Asia at significant frequencies.
E-M243 is common among Afro-Asiatic speakers in the Near East and North Africa as well as among some Nilo-Saharan and Niger–Congo speakers in North East Africa and Sudan.
E-M243 is far less common in West, Central, and Southern Africa, though it has been observed among some Khoisan speakers and among Niger–Congo speakers in Senegambia, Guinea-Bissau, Burkina Faso, Ghana, Gabon, the Democratic Republic of the Congo, Rwanda, Namibia, and South Africa.
Single nucleotide polymorphism
Filed under: Uncategorized